Introduction
In the article series Efficient Realtime Signal Processing C++ Templates, we demonstrate highly efficient and manageable implementation methods and libraries for complex real-time signal processing and machine learning pipelines. This series provides an application example showing how these techniques and libraries support flexible, reliable, and maintainable implementations for electrocardiogram (ECG) signal conditioning as well as QRS complex detection, feature extraction, and classification. The first installment focuses on signal conditioning. See [1] and [2] for more signal processing detail underlying this implementation.
The ECG is a set of electrical signals derived from electrodes placed on the thorax or limbs. These signals represent the superposition of cellular electrical action potentials originating from pacemaker cells in the heart, which propagate through heart tissue and the body to reach surface electrodes. By analyzing features of these signals, clinicians and electrophysiologists gain insight into heart health and can detect irregular heart rhythms such as premature beats or atrial fibrillation. These signals are even more valuable when continuously and automatically monitored by automated machine learning or artificial intelligence algorithms – whose practical, efficient, and state-of-the-art realtime implementation is an area of focus for this series of articles.
An ECG waveform is a differential signal typically derived from 2 to 10 electrodes placed at standard points on the body. The time-varying differential voltage between any two of these electrodes is referred to as an ECG lead or vector. There can be any number of ECG leads computed. Wearables or Band-Aid style ECG patches often have 2 electrodes. In vital signs monitors a driven electrode often serves as a ground reference for 2 to 9 other electrodes to support 3 to 12 lead ECG systems. In the common and standard 12-lead ECG, the leads are labeled I, II, III, AVR, AVL, AVF, V1, V2, V3, V4, V5, and V6. The following code example shows how to compute them from 9 single-ended electrode inputs that use a right leg electrode as the driven ground reference.
void Leads(VEC_LEADS_12& out, const VEC_ELECTRODES& in)
{
// in[0] is right leg ground reference for other single-ended electrodes.
T LL = in[1]; // Left leg.
T RA = in[2]; // Right arm.
T LA = in[3]; // Left arm.
T wilsonCentral = (LA + RA + LL) / 3;
out[0] = LA - RA; // I
out[1] = LL - RA; // II
out[2] = LL - LA; // III
out[3] = 3 * (RA - wilsonCentral) / 2; // aVR
out[4] = 3 * (LA - wilsonCentral) / 2; // aVL
out[5] = 3 * (LL - wilsonCentral) / 2; // aVF
out[6] = in[4] - wilsonCentral; // V1
out[7] = in[5] - wilsonCentral; // V2
out[8] = in[6] - wilsonCentral; // V3
out[9] = in[7] - wilsonCentral; // V4
out[10] = in[8] - wilsonCentral; // V5
out[11] = in[9] - wilsonCentral; // V6
}A variable called wilsonCentral represents the “wilson central terminal” which is an average of the left arm (LA), right arm (RA), and left leg (LL) electrodes that provides a central body reference to compute the augmented limb leads (aVR, aVL, aVF) and the chest (precordial) leads (V1-V6).
Signal Conditioning C++ Template
ECG leads are prone to various types of noise and artifact that can affect their interpretation, making signal conditioning an essential first stage of processing. Typically, there are four main types of noise or artifact with corresponding filters:
Baseline Wander – low frequency changes or drift in the signal often from patient movement, such as impedance changes due to respiration. A high-pass filter with a cutoff frequency of around 0.5–1 Hz is often used to remove. There are also many other removal methods, such as Wavelet denoising or estimating the baseline drift using a polynomial fit and then subtracting.
Muscle Noise – a designation for any higher frequency noise where muscle activation potentials are significant contributors. However, this noise can come from many other environmental sources, such as electrocautery instruments. A lowpass filter with a cutoff frequency of around 100 Hz is commonly used to mitigate.
Powerline Noise – AC powerline interference of typically (50/60 Hz) is ubiquitous interference in any electronics with connection to a wall socket. A notch filter typically removes it, but because of the challenge in creating a narrow notch via linear time invariant methods, adaptive techniques are often used that adjust their coefficients in real-time to more precisely eliminate a single frequency while also minimizing computation and filter I/O delay. The implementation below employs a classic option described in [1] that is widely used commercially.
Pacemaker Spikes – Pacemakers generate brief, localized electrical impulses to stimulate the heart, compensating for abnormalities in the natural cardiac pacemaker cells or the electrical conduction pathways that coordinate the contraction and relaxation of the heart. These impulses do not resemble natural ECG waves but instead appear as sharp spikes lasting a few milliseconds, with a height between 2 and 20 mV, which can be either positive or negative depending on the lead placement and impulse orientation. The position of the spike in the ECG varies according to the pacing mode:
- Atrial pacing: The spike appears before the P wave.
- Ventricular pacing: The spike appears just before the QRS complex.
- Dual-chamber pacing: Spikes precede both the P wave and the QRS complex.
Compared to older pacemakers, modern pacemakers generate less artifact but can also cause more complex signal dynamics. Rate adaptation pacing modes synchronize better with the heart’s natural speed ups and slowdowns with changes in metabolic demands, caused by exertion for example, which results in a more natural ECG. Because heartbeat time intervals and strength are both modulated beat-to-beat by various physiological influences, exact periodic pacing, for example, is very unnatural. In-demand pacing modes monitor natural pacing, where the pacemaker turns itself on only when natural pacing fails and turns itself off when natural pacing is adequate.
Dealing with pacemakers can be a significant signal processing challenge, but mathematical morphology filtering [TODO: provide link to future mathematical morphology filtering blog entry] is an elegant and efficient nonlinear approach for removing or identifying pacemaker spikes of any type within the ECG. This method, which requires no multiplications and allows for fast implementation, is well-suited for use in wearables that require long battery life.
The following template implements a complete, efficient, high quality, flexible ECG signal conditioning frontend with configurable bandwidths, powerline frequency, enable/disable of each filter stage, etc. The frontend takes a vector of multiple ECG leads as a template parameter and can thus process any number of ECG leads in parallel.
#include <esf/stddef.h>
#include <esf/dsp/filter.hpp>
#include <esf/dsp/filterBoxcar.hpp>
#include <esf/dsp/filterSharpenPoly10.hpp>
#include <esf/dsp/filterSimpleHighpass.hpp>
#include <esf/dsp/filterPowerline.hpp>
NAMESPACE(ESF)
NAMESPACE(ECG)
template<typename TVec>
class SignalConditioner {
public:
// For a bandwidth configurable Poly10 lowpass filter.
static constexpr int N_MAX_LP = 64; // Maximum boxcar filter length
static constexpr int N_DEFAULT_LP = 28; // Must be <= to N_MAX_LP
using T = typename TVec::Scalar;
static constexpr T SAMPLE_FREQ = static_cast<T>(2000.0); // Default value.
// Default powerline filter configuration.
static constexpr T PWR_DELTA = static_cast<T>(0.00001); // Step size for adaptation.
static constexpr T PWR_LINE_FREQ = static_cast<T>(60.0); // Default freq.
// Baseline wander removal filter cutoff default frequency.
static constexpr T CUTOFF_HIGH_1_0_HZ = static_cast<T>(0.996863331833438);
using FILTER = DSP::FilterBoxcarN<TVec, N_MAX_LP>;
using FILT_LOWPASS = DSP::FilterSharpenPoly10<FILTER, N_MAX_LP>;
using FILT_LOWPASS_CFG = FILTER::Config;
using FILT_POWERLINE = DSP::FilterPowerline<TVec>;
using FILT_POWERLINE_CFG = typename FILT_POWERLINE::Config;
using FILT_BASELINE = DSP::FilterSimpleHighpass<TVec>;
using FILT_BASELINE_CFG = typename FILT_BASELINE::Config;
// Put ALL configuration here.
struct Config
{
Config(int nLp = N_DEFAULT_LP, T stateLp = static_cast<T>(0.0),
T baseCutoff = CUTOFF_HIGH_1_0_HZ,
T stateBase = static_cast<T>(0.0), T delta = PWR_DELTA,
T fs = SAMPLE_FREQ, T fn = PWR_LINE_FREQ,
bool dl = true, bool dh = false, bool dp = false)
: lowpass(static_cast<int>(nLp*SAMPLE_FREQ/fs), stateLp),
baseline(baseCutoff*SAMPLE_FREQ/fs, stateBase),
powerline(delta, fs, fn),
doLowpass(dl),
doBaseline(dh),
doPowerline(dp)
{
}
Config(const Config &c) = default;
FILT_LOWPASS_CFG lowpass; // Lowpass filter configuration.
FILT_POWERLINE_CFG powerline; // 50 /60 Hz powerline filter configuration.
FILT_BASELINE_CFG baseline; // Baseline wander high pass filter configuration.
bool doLowpass; // Run lowpass filter?
bool doBaseline; // Run highpass baseline wander removal filter?
bool doPowerline; // Run powerline notch filter?
bool operator==(const Config &rhs) const
{
return (lowpass == rhs.lowpass &&
powerline == rhs.powerline &&
baseline == rhs.baseline &&
doLowpass == rhs.doLowpass &&
doBaseline == rhs.doBaseline &&
doPowerline == rhs.doPowerline);
}
};
SignalConditioner(const Config &cfg)
: cfg(cfg), filtBaseline(cfg.baseline),
filtPowerline(cfg.powerline), filtLowpass(cfg.lowpass)
{}
virtual ~SignalConditioner() = default;
bool Init(const Config &c)
{
cfg = c;
return (filtLowpass.Init(cfg.lowpass) &&
filtBaseline.Init(cfg.baseline) &&
filtPowerline.Init(cfg.powerline));
}
bool Get(Config *pC) const { *pC = cfg; return true; }
T Delay() const
{
T delay = 0.0;
if (cfg.doPowerline)
delay += filtPowerline.Delay();
if (cfg.doBaseline)
delay += filtBaseline.Delay();
if (cfg.doLowpass)
delay += filtLowpass.Delay();
return delay;
}
// The main filtering operator.
int operator()(TVec &out, const TVec &in)
{
// Always run the baseline filter whether it is externally enabled
// or not. Decide later to use the result if the filter is enabled,
// else do not use it. This filter's transient response may be
// several seconds long, which would show up on a display
// as the filter is dynamically enabled/disabled. Having the
// internal state precomputed this way supports dynamic enable/disable
// of this filter without seeing its transient response.
filtBaseline(state[0], in);
// If the baseline filter is not enabled, replace the filter result.
if (!cfg.doBaseline)
{
state[0] = in;
}
// If low-pass is enabled,
if (cfg.doLowpass)
{
filtLowpass(state[1], state[0]);
}
else // Bypass the filter.
{
state[1] = state[0];
}
// Always run the powerline notch filter and store the result
// whether it is externally enabled or not. Decide later to use the
// result if the filter is enabled, else do not use it. This filter's
// transient response may be several seconds long, which would show up on
// a display as the filter is dynamically enabled/disabled. Having the
// output precomputed this way supports dynamic enable/disable of this
// filter without seeing its transient response.
filtPowerline(out, state[1]);
// If the powerline filter is not enabled, replace the computed result.
if (!cfg.doPowerline)
{
out = state[1];
}
return 1;
}
private:
Config cfg;
FILT_LOWPASS filtLowpass;
FILT_BASELINE filtBaseline;
FILT_POWERLINE filtPowerline;
// Internal state.
TVec state[2];
};
NAMESPACE_END(ECG)
NAMESPACE_END(ESF)
Note that implementation consists of selecting and configuring the desired filter types from the DSP template library and then writing a short loop to run them. The lowpass filter is the computationally efficient sharpened filter introduced in the article, Efficient Realtime Signal Processing C++ Templates and described in [2], and the powerline filter is an improved version of an efficient classic adaptive filter method to remove powerline noise [1] that removes the first and next several powerline frequency harmonics. The highpass filter in this example is a single-pole, single-zero recursive filter that has low real-time I/O delay (supports total signal conditioning I/O delay < 50 ms). However, unlike other component filters in the conditioning, this filter has nonlinear phase where higher frequencies have low I/O delay and lower frequencies longer delay. If bandwidth is set too high, it would cause the frontend to fail performance standards such as EC-13. However, if higher I/O delay is tolerable, it would be easy to replace this highpass with the optimal (highest cutoff while still passing EC-13 pulse distortion tests) linear phase option introduced in [2].
Default configuration parameters for the above example include a 2 kHz sampling frequency that is on the high-end side for ECG. However, all parameters are scaled at the top level without any underlying filter changes to support configuration of any sampling frequency. The template parameter is a vector type representing the number of leads, so this single template can be instanced to process any number of ECG leads – 1, 3, 5, 12, etc.
Implementation Example
Using PyBind11 discussed in article Python Binding of Realtime C++, we can easily instance ECG signal conditioning frontends for different numbers leads and expose them to Python. First instance the ECG::SignalConditioner class using the following template function,
#include <pybind11/eigen.h>
#include <pybind11/numpy.h>
#include <pybind11/pybind11.h>
#include <pybind11/stl.h>
#include <esf/math/vector.hpp>
#include <esf/math/vectorN.hpp>
#include <esf/biomed/electrophysiology/ecg/SignalConditioner.hpp>
namespace py = pybind11;
namespace ESF {
namespace ECG {
// Use a separate function to bind the powerline filter config class for every
// instance of template<int TNumLeads> void SignalConditionPyBind11(). Not
// sure why, but Python thinks the name is registered twice. The number of leads
// is in the name, so one would think this is sufficient. It is sufficient
// for the other filters composing the signal conditioner. The only difference
// between the powerline and other filters is that the powerline Config struct
// is defined in a PowerlineKernel class that is the same regardless of the number
// of leads.
void RegisterConfigPowerline(py::module& m) {
// Use 1 lead to get the definition. It does not matter how many leads are used.
using ConfigPowerline = ECG::SignalConditioner<Math::VectorN<double, 1>>::FILT_POWERLINE_CFG;
// Binding the ConfigPowerline class once for all template instances.
py::class_<ConfigPowerline>(m, "ConfigPowerline", "Configuration for ECG powerline notch filter")
.def(py::init<>()) // Default constructor
.def(py::init<const ConfigPowerline&>()) // Copy constructor
.def_readwrite("delta", &ConfigPowerline::delta) // Step size for adaptive filter.
.def_readwrite("fs", &ConfigPowerline::fs) // Sampling frequency.
.def_readwrite("fn", &ConfigPowerline::fn); // Frequency to remove.
}
// Bind the SignalConditioner class for a specific number of leads.
template<int TNumLeads>
void SignalConditionPyBind11(py::module& m, const std::string& name) {
using VEC_LEADS = Math::Vector<NUMBER>;
using VEC_LEADS_N = Math::VectorN<NUMBER, TNumLeads>;
// Signal conditioning for ECG differential signals.
using Class = ECG::SignalConditioner<VEC_LEADS_N>;
using Config = Class::Config;
using ConfigLowpass = Class::FILT_LOWPASS_CFG;
using ConfigBaseline = Class::FILT_BASELINE_CFG; // Baseline wander removal filter configuration.
using ConfigPowerline = Class::FILT_POWERLINE_CFG;
// Bind the Lowpass filter config class.
py::class_<ConfigLowpass>(m, (name + "ConfigLowpass").c_str(), "Configuration for ECG lowpass filter")
.def(py::init<>()) // Default constructor
.def(py::init<const ConfigLowpass&>()) // Copy constructor
.def_readwrite("N", &ConfigLowpass::N)
.def_readwrite("val", &ConfigLowpass::val);
// Bind the baseline wander removal filter config class.
py::class_<ConfigBaseline>(m, (name + "ConfigBaseline").c_str(), "Configuration for ECG baseline wander removal filter")
.def(py::init<>()) // Default constructor
.def(py::init<const ConfigBaseline&>()) // Copy constructor.
.def_readwrite("alpha", &ConfigBaseline::alpha) // Feedback coefficient.
.def_readwrite("stateInit", &ConfigBaseline::stateInit);
// Bind the lowpass filter config class
py::class_<Config>(m, (name + "Config").c_str(), "Configuration for ECG SignalConditioner")
.def(py::init<>()) // Default constructor
.def(py::init<const Config&>()) // Copy constructor
.def_readwrite("lowpass", &Config::lowpass) // ConfigLowpass member
.def_readwrite("baseline", &Config::baseline) // ConfigBaseline member
.def_readwrite("powerline", &Config::powerline)
.def_readwrite("doLowpass", &Config::doLowpass) // Run lowpass filter?
.def_readwrite("doBaseline", &Config::doBaseline) // Run highpass baseline wander removal filter?
.def_readwrite("doPowerline", &Config::doPowerline); // Run powerline notch filter?
// Bind the main SignalConditioner class
py::class_<Class>(m, (name).c_str(), "EcgSignalConditioner class")
.def(py::init<const Config&>(), "Construct with Config object")
.def("Init", py::overload_cast<const Config&>(&Class::Init), "Initialize with Config")
.def("Get", &Class::Get, "Get Config parameters")
.def("__call__", [](Class& instance, const py::array_t<NUMBER>& in) {
auto in_buf = in.request();
// If the input is a scalar, return a scalar.
if (in_buf.ndim == 0) {
if (TNumLeads == 1) {
// The input/output vectors are views on the same memory to avoid data copies.
const VEC_LEADS_N vecIn(static_cast<NUMBER*>(in_buf.ptr));
py::array_t<NUMBER> out_array = py::array_t<NUMBER>(1);
auto out_buf = out_array.request();
VEC_LEADS_N vecOut(static_cast<NUMBER*>(out_buf.ptr));
py::gil_scoped_release release;
int result = instance(vecOut, vecIn);
py::gil_scoped_acquire acquire;
return out_array; // Return as a NumPy array
}
else {
throw std::runtime_error("If input is a scalar then TNumLeads=" +
std::to_string(TNumLeads) + " must be 1.");
}
// If the input is a 1D array,
} else if (in_buf.ndim == 1) {
// If the number of leads equals the size of the array, assume that this
// is single sample of all leads, and process it as a vector.
if (TNumLeads == in_buf.shape[0]) {
const VEC_LEADS_N vecIn(static_cast<NUMBER*>(in_buf.ptr));
py::array_t<NUMBER> out_array({ in_buf.shape[0] });
auto out_buf = out_array.request();
VEC_LEADS_N vecOut(static_cast<NUMBER *>(out_buf.ptr));
py::gil_scoped_release release;
int result = instance(vecOut, vecIn);
py::gil_scoped_acquire acquire;
return out_array;
}
// If this is a 1 lead ECG, assume the input is single samples and process them.
else if (TNumLeads == 1) {
py::array_t<NUMBER> out_array({ in_buf.shape[0] });
NUMBER* pOutBuf = static_cast<NUMBER*>(out_array.request().ptr);
NUMBER* pInBuf = static_cast<NUMBER*>(in_buf.ptr);
VEC_LEADS_N vecIn, vecOut;
py::gil_scoped_release release;
for (int i = 0; i < in_buf.shape[0]; i++, pInBuf++, pOutBuf++) {
vecIn.Init(pInBuf); // Create a view on external memory.
vecOut.Init(pOutBuf);
int result = instance(vecOut, vecIn);
}
py::gil_scoped_acquire acquire;
return out_array;
// Leads and samples do not match.
} else {
throw std::runtime_error("If TNumLeads=" + std::to_string(TNumLeads) +
" > 1, then it must match the size of a 1D array input.");
}
// Process a 2D array, and make sure 1 of the dimensions matches TNumLeads.
} else if (in_buf.ndim == 2) {
// If the number of leads equals the number of columns, process each row at a time.
// Numpy is row-major, so we only support leads as columns to support viewing contiguous
// memory without copying and to better support caching the sample vector of leads.
if (TNumLeads == in_buf.shape[1]) {
py::array_t<NUMBER> out_array({ in_buf.shape[0], in_buf.shape[1] });
auto out_buf = out_array.request();
VEC_LEADS_N vecIn, vecOut;
size_t rowStride = in_buf.strides[0];
py::gil_scoped_release release;
for (int i = 0; i < in_buf.shape[0]; i++) {
NUMBER* pInRow = (NUMBER*)(static_cast<char*>(in_buf.ptr) + i * rowStride);
NUMBER* pOutRow = (NUMBER*)(static_cast<char*>(out_buf.ptr) + i * rowStride);
vecIn.Init(pInRow);
vecOut.Init(pOutRow);
int result = instance(vecOut, vecIn);
}
py::gil_scoped_acquire acquire;
return out_array;
}
else {
throw std::runtime_error("For a 2D array, the rows=" + std::to_string(TNumLeads) +
" must equal TNumLeads=" + std::to_string(TNumLeads) + ".");
}
} else {
throw std::runtime_error("The number of input array dimensions, ndim=" +
std::to_string(in_buf.ndim) + "can at most be 2D");
}
}, py::return_value_policy::reference_internal, "Call operator() to process input and output arrays and return the integer result.")
.def("Delay", &Class::Delay, "Get the I/O delay in sample periods");
}
} // namespace ECG
} // namespace ESFData moves between NumPy arrays and the filter’s vector inputs/outputs without memory allocation or data copies because vecIn.Init(...) and vecOut.Init(...) set up a view for these vectors to directly use the same NumPy array memory. The following code is then used to build the Python module that includes several ECG signal conditioning frontends with different numbers of leads:
void Ecg(py::module&);
PYBIND11_MODULE(electrophysiology, m) {
m.doc() = "Electrophysiology signal processing module";
// Initialize the ECG submodule
py::module ecgModule = m.def_submodule("ecg", "ECG signal processing submodule");
Ecg(ecgModule);
}
void Ecg(py::module& m) {
// See the comments related to this function to understand why it is needed.
ECG::RegisterConfigPowerline(m);
// Signal conditioners for different numbers of ECG leads.
ECG::SignalConditionPyBind11<1>(m, "EcgSignalConditioner1");
ECG::SignalConditionPyBind11<2>(m, "EcgSignalConditioner2");
ECG::SignalConditionPyBind11<3>(m, "EcgSignalConditioner3");
ECG::SignalConditionPyBind11<5>(m, "EcgSignalConditioner5");
ECG::SignalConditionPyBind11<12>(m, "EcgSignalConditioner12");
// QRS detection, feature extraction, classification, etc.
}The following example Python code downloads a 2-lead ECG signal from Physionet.org, filters it with one line of Python using the 2-lead external realtime C++ filter implementation, and then plots one of the leads with its filtered version shifted by the I/O delay of the filter returned by the Delay() operator. Because of its real-time design, the external C++ signal conditioner maintains state efficiently (minimal copies and zero memory allocation) via its internal circular buffers and the external code can iteratively send it samples forever in whatever chunks at time that it wishes – 1, 2, 10, 100, or N.
import matplotlib.pyplot as plt
import wfdb
import sigSmart.electrophysiology as electrophys
# Fetch the record and annotation from PhysioNet
record_name = '100'
record = wfdb.rdrecord(record_name, pn_dir='mitdb')
signal = record.p_signal
# Configure to match the ECG signal sampling frequency.
cfg = electrophys.ecg.SignalConditioner2Config()
cfg.lowpass.N = 4 # Bandwidth selection; larger -> lower bandwidth.
signalCondition = electrophys.ecg.SignalConditioner2(cfg)
# Signal condition using the external C++ code.
sig_filt = signalCondition(signal)
# Plot original ECG and filtered version on the same graph; correct for filter delay.
lead_idx = 0
time = [i / record.fs for i in range(len(signal[:,lead_idx]))]
delay = int(signalCondition.Delay())
plt.figure(figsize=(15, 5))
label = 'ECG Lead '+str(lead_idx)
plt.plot(time, signal[:, lead_idx], label=label)
plt.plot(time[:-delay], sig_filt[delay:,lead_idx], label=label + ' Filtered')
plt.xlabel('Time (s)')
plt.ylabel('Amplitude (mV)')
plt.title(f'WFDB Record {record_name}')
plt.legend()
plt.grid()
plt.show()An example output from the Python code is:
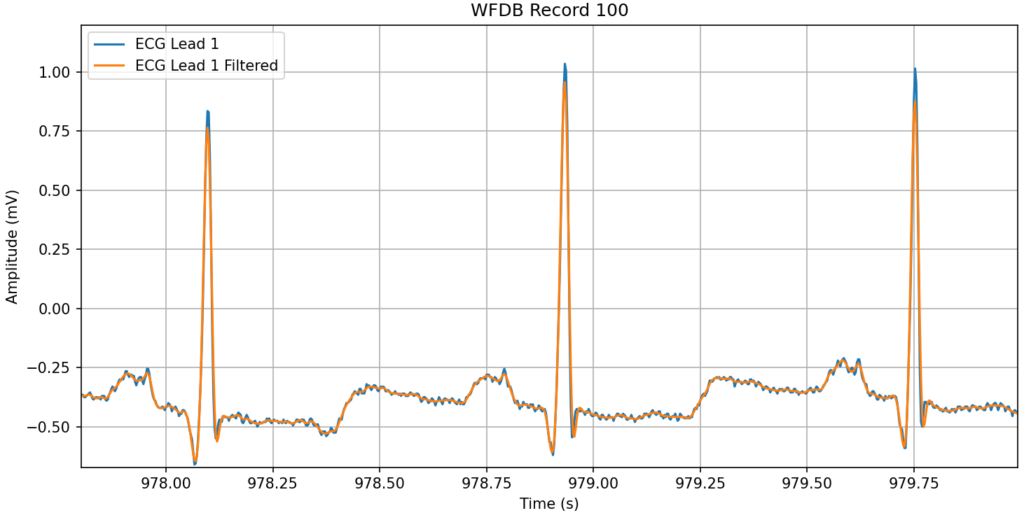
Bibliography
[1] M.L. Ahlstrom and W.J. TOMPKINS. “Digital filters for real-time ECG signal processing using microcomputers“, IEEE Trans Biomed Eng, Vol BME-32, No. 9, Sept. 1985, pp 708-713
[2] Alexander Holland, [Online]. Available: Signal Conditioning with Almost No Multiplications via Filter Sharpening | IEEE Conference Publication | IEEE Xplore
